Recombinant DNA Technology
Let us imagine that you are going back to the early 1970s. You had to completely depend on insulin harvested from the pancreases of slaughtered pigs and cows for your survival if you were diagnosed with Type 1 Diabetes. It took tons of animal pancreases to produce just a few ounces of insulin. As it was not human insulin, it was expensive, inconsistent, and many people developed allergic reactions to it. It was barely a life-saving treatment for the people who were suffering from diabetes. Then things changed with the dawn of Recombinant DNA Technology.
There began a quiet revolution in the laboratories that altered our relationship with the biological world. Researchers started editing nature’s blueprint – DNA, instead of just observing it. They figured out the method to take out the human gene responsible for producing insulin and insert it into a bacterium. These bacteria were tricked into becoming microscopic factories, churning out pure human insulin.
Recombinant DNA Technology is becoming the foundation of modern biology, medicine, and agriculture for all life science students worldwide. In this article, let us dive into the learning of the cut and paste method for the code of life.
What is Recombinant DNA Technology?
Life is defined by the information stored in the DNA. For billions of years, it was believed that the genetic information could generally be shared within a species. A potato plant could not share genes with a fish.
But Recombinant DNA Technology breaks that rule!
So you can ask now, What is Recombinant DNA Technology? What it does? And many more questions may arise in your mind..
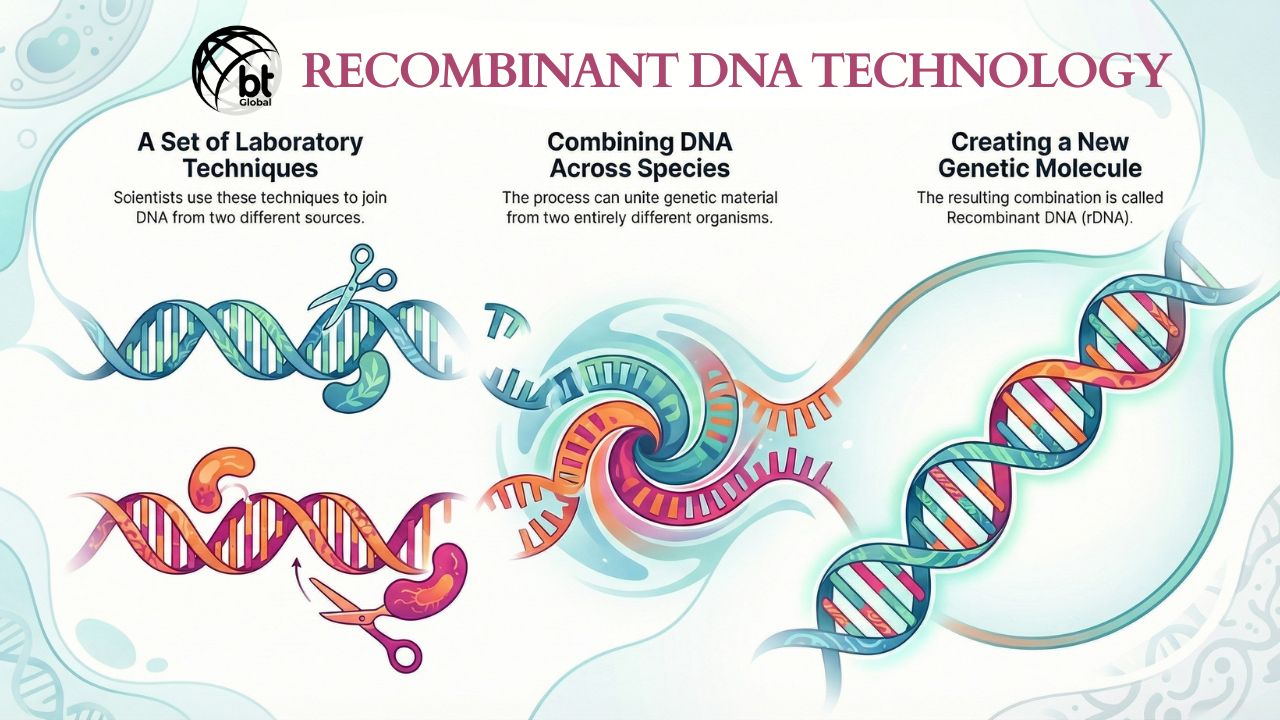
Recombinant DNA Technology is a set of laboratory techniques that allows researchers to combine DNA from two different organisms of entirely different species to create a new genetic combination. The resulting molecule will be termed as Recombinant DNA (rDNA).
Consider it like editing a videotape. Think of the DNA of the host as the original movie, and the gene of interest as a clip from another film that you wish to insert into it. You will need specific tools to cut the original videotape, create a slot for the new clip, and tape it all together so it plays smoothly. When this edited new videotape is placed inside a living cell, the cell starts reading the new instructions. With the latest instructions, the cell begins to produce a specific protein. This is called a Recombinant Protein.
The Essential Enzymes
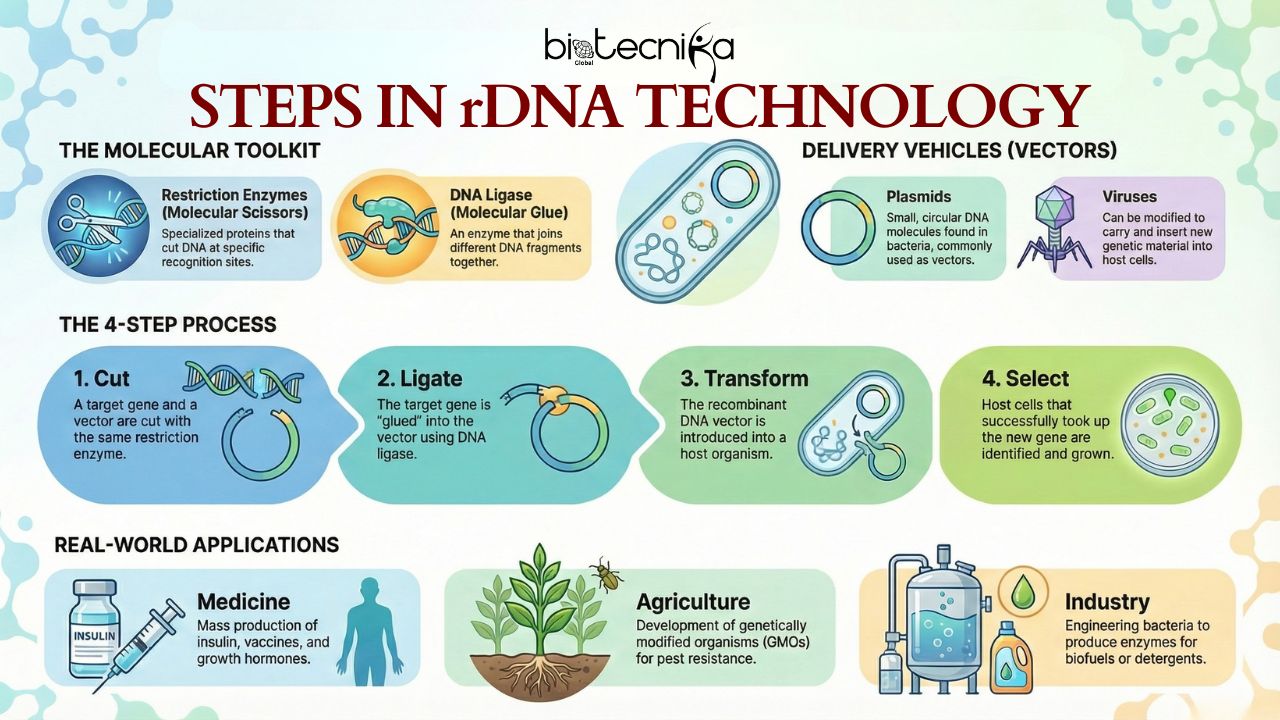
We will need a saw and glue before we try to build furniture. In genetic engineering, the tools are the enzymes. You cannot master Recombinant DNA Technology without knowing these players. Restriction enzymes and ligases are the most important enzymes used in Recombinant DNA Technology.
1. Molecular Scissors
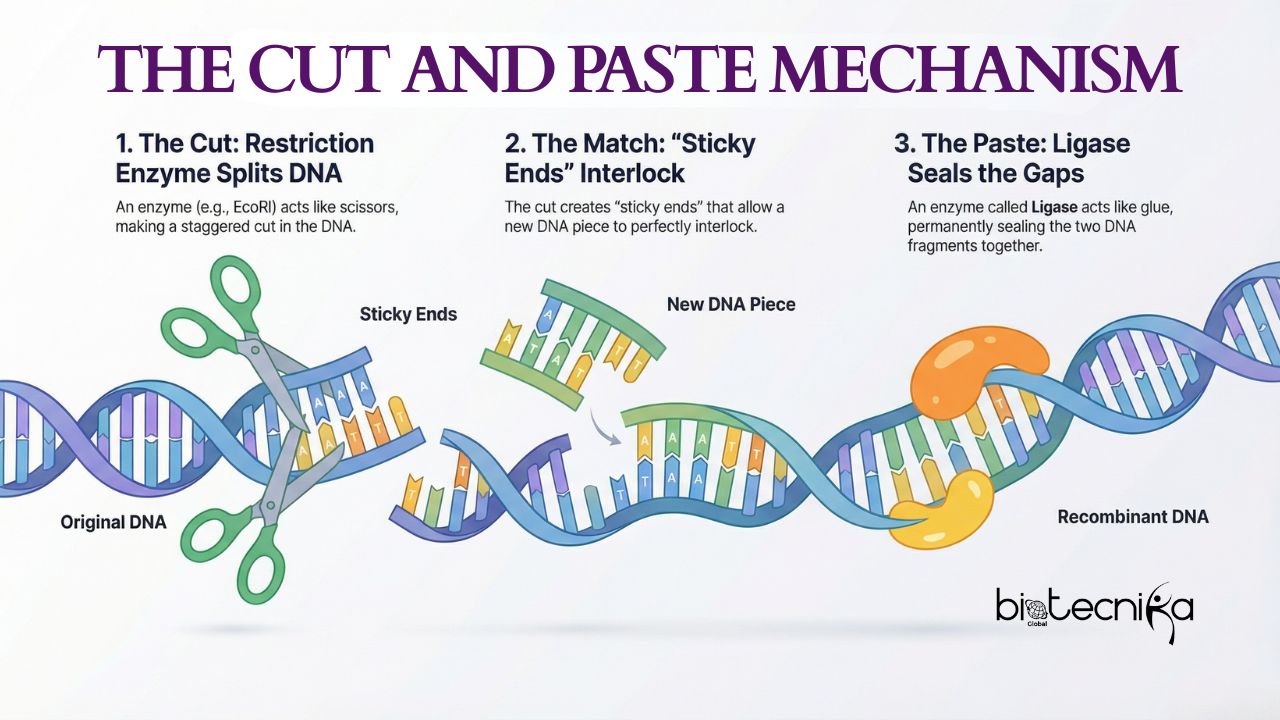
- DNA is a very long molecule. We need to cut it out to work with a specific gene. Restriction enzymes, or Restriction endonucleases, are used for cutting. These restriction enzymes are naturally found in bacteria.
- They act as a crude immune system, chopping off the viral DNA invading a bacterium. They do not cut randomly. They scan the DNA and look for specific sequences (usually 4 to 8 base pairs long). These sequences are known as “Recognition sites”.
- Many restriction enzymes make staggered cuts, leaving short, single-stranded overhangs at the ends. These are called “Sticky ends.” And some restriction enzymes create “Blunt ends.” These blunt ends are created when restriction enzymes make a straight cut across both strands. Blunt ends lack any single-stranded overhangs.
- Common Restriction Enzymes and Their Restriction Sites
| Enzyme Name | Source Organism | Recognition Sequence (The cut is usually between the first two bases) | Type of Ends Produced |
| EcoRI | Escherichia coli | GAATTC | Sticky Ends |
| BamHI | Bacillus amyloliquefaciens | GGATCC | Sticky Ends |
| HindIII | Haemophilus influenzae | AAGCTT | Sticky Ends |
| SmaI | Serratia marcescens | CCCGGG | Blunt Ends (Straight cut, no overhang) |
Nomenclature of Restriction Enzymes
The naming system of restriction enzymes follows a standard convention proposed by Smith and Nathans. The names are derived from the organism from which they are isolated.
Here is the breakdown of the rules using the famous enzyme EcoRI as an example:
- Rules of Nomenclature
- First Letter – Genus: The first letter of the name of a restriction enzyme comes from the Genus of the organism. It should always be capitalized and written in italics.
E – Escherichia
2. Second and Third Letter – Species: The following two letters come from the species of the organism. It should always be in lowercase letters and italicized.
co – coli
3. Fourth Letter – Strain: The fourth letter indicates the specific strain type of the organism. This should not be italicized.
R – Strain RY13
4. Roman Numerals – Order: The Roman numeral at the end indicates the order in which the enzyme was isolated from the specific strain of the organism.
I – First enzyme isolated from a particular strain.
2. Vehicles
- You have isolated the gene of interest. Now, how do you get it into a host cell and ensure it stays there and replicates? In biology, a delivery vehicle is called a vector. The most common vectors used in research labs are plasmids.
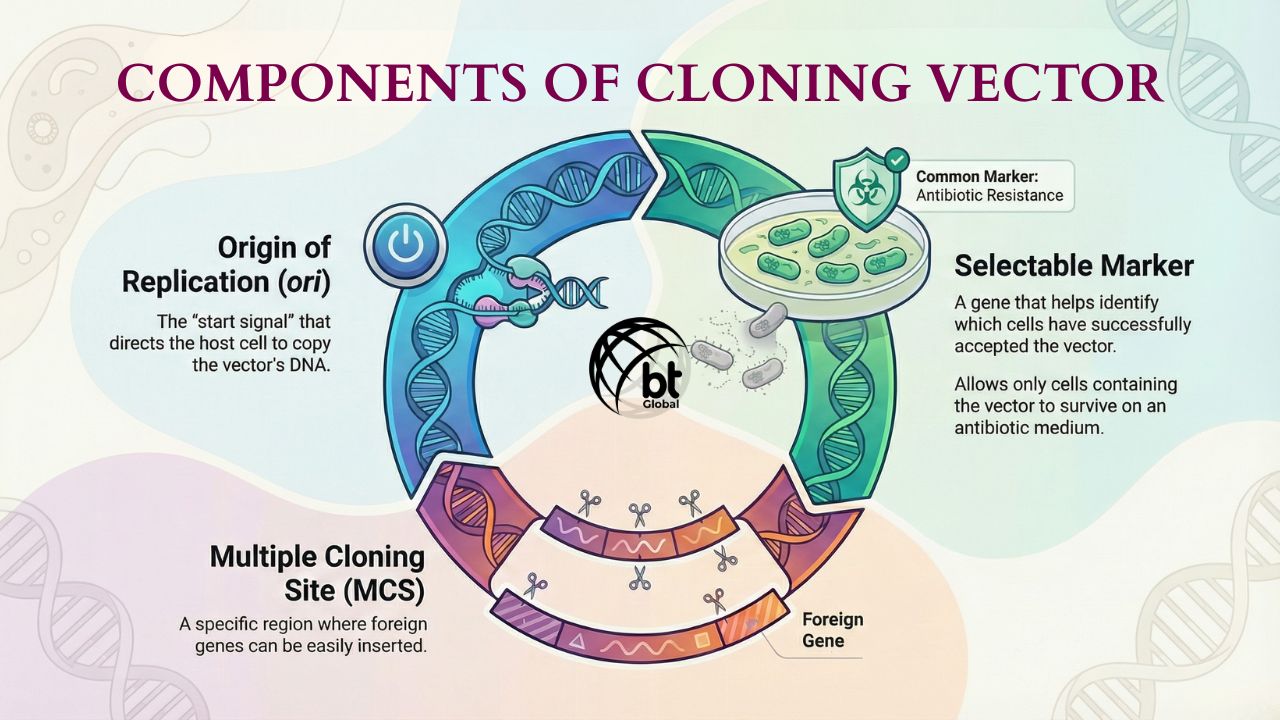
- Plasmids are small, covalently closed, circular DNA found inside bacteria. They are separate from the main chromosome. They are also self-replicating. The following things make a plasmid a good candidate for a cloning vector,
- Origin of Replication (ori): They are short sequences in the plasmid that give a starting signal for the replication process to begin.
- Selectable Marker: It helps tag a gene with antibiotic resistance. For example, the Ampicillin resistance gene allows us to identify which bacteria accepted the plasmid.
- Multiple Cloning Site (MCS): This is where the gene sequences are rich in recognition sites for various restriction enzymes. In these sites, you can easily insert the gene of interest.
3. Molecular Glue
- Once you have the DNA of the host and the gene of interest with a vector, the sticky ends will find each other due to base pairing ( A double bond T and C triple bond G). However, there will be some gaps in the backbone of the DNA ladder.
- DNA Ligases are enzymes used to seal these gaps. They form phosphodiester bonds, making the new recombinant DNA a solid, continuous piece of DNA.
5-Step Process for making Recombinant DNA
As we now have the tools and the vehicle, let us walk through the actual workflow for making Recombinant DNA. This is the bread and butter of a molecular biology laboratory.
Step 1: Isolation of Genetic Material
At first, you need the DNA. The cells of the organism containing the gene of interest and the bacterial cells containing the plasmid vector should be lysed. The DNA is isolated using the standard alkaline lysis method to separate it from proteins and lipids.
Step 2: Cutting (Restriction Digestion)
- Precision happens here. The gene of interest and the plasmid vector are put into separate test tubes. Then, a restriction enzyme is added to both tubes (e.g., EcoRI)
- The enzyme cuts the DNA from the desired organism into many fragments. One of those small fragments contains the gene of interest.
- The same enzyme also cuts the plasmid vector and opens it into a linear strand.
- As the same enzyme cuts both the gene of interest and the plasmid vector, there will be matching sticky ends.
Step 3: Joining (Ligation)
The gene of interest and the plasmid vector are put together in the same tube. Then, DNA Ligase is added to the tube. The sticky ends of the gene of interest will base-pair with the sticky ends of the plasmid vector. The ligase enzyme will seal to create a circular plasmid containing the host genome. This is the Recombinant DNA molecule.
Step 4: Transformation (Getting it into the host)
- The rDNA is still just in a test tube. It needs to be inside a living host to work. Transformation is the process of introducing a foreign gene into the host cells.
- The host organism doesn’t readily take up the naked DNA. It should be made ready to accept a foreign gene. This is called the process of making them competent. They are made competent by the treatment of calcium chloride and a sudden heat shock. The cell wall of the organism temporarily opens by creating pores that allow the rDNA to slip inside them.
Step 5: Screening and Selection (Finding the Needle in the Haystack)
- We get a tube of bacteria after transformation. We will not know which bacteria took up the rDNA and which didn’t, as they are invisible to the eye. So, how do we find the right ones?
- Selectable Markers, such as antibiotic resistance genes, are used to identify rDNA.
- Bacteria are grown on an agar plate containing an antibiotic such as ampicillin.
- Bacteria that do not take up the rDNA will die because they are not resistant to the given antibiotic.
- Bacteria that have taken up the rDNA will survive because they are resistant to the given antibiotic and will grow into colonies.
- Blue-white screening is one of the most common methods used to identify them.
Blue-White Screening
The insertion site on the plasmid is located within a gene called lacZ. If your gene inserts correctly, it breaks the lacZ gene.
- If the lacZ is intact (empty plasmid), the bacteria turn blue on special media.
- If the lacZ is broken by your inserted gene (recombinant plasmid), the bacteria remain white.
- Therefore, the white colonies growing on the antibiotic plate are the ones you want!
Applications of Recombinant DNA Technology
We don’t just do this because it’s cool science. Many Global problems can be solved using this genomics technology. The ability to produce recombinant proteins has changed the world in terms of agriculture, the environment, industry, and medicine. Some of the applications are given below,
|
Field |
Application Example |
Impact |
| Medicine | Human Insulin Production (Humulin) | Replaced animal insulin; purer, safer supply for diabetics. |
| Medicine | Tissue Plasminogen Activator (tPA) | Dissolves blood clots; used to treat heart attacks and strokes. |
| Vaccines | Hepatitis B Vaccine | Produced in yeast cells containing the viral gene; safer than traditional vaccines. |
| Agriculture | Bt Cotton / Bt Corn | Plants engineered with a bacterial gene to produce their own pesticide, reducing chemical spraying. Many Plant Biology Milestones have been created. |
| Agriculture | Golden Rice | Rice engineered to produce Vitamin A, combating blindness in developing nations. |
| Industry | Detergent Enzymes | Proteases and lipases produced by bacteria help clean clothes at lower temperatures. |
Recombinant DNA Technology is more than just a chapter in a textbook. It is defining the technology of our biological age. It is more into designing what organisms can be than describing what organisms are. You will definitely encounter these concepts as you move forward in your life science studies. The ability to cut, paste, and read DNA will be the most fundamental skill, whether you are studying cancer genetics, researching novel antibiotics, or developing drought-resistant crops. The revolution that started with insulin extends to the next generations, too.